Epigenetic Regulation of Retinal Development
Previous work in our laboratory focused on the regulation of proliferation and differentiation in retinal progenitor cells (retinoblasts) and within the stem cell containing ciliary marginal zone (CMZ) of the retina. We demonstrated that de novo purine synthesis regulates retinoblast proliferation, wherein an ATP-dependent pathway modulates cell cycle kinetics in proliferative retinoblasts (Ng et al., 2009). We identified a mutation in cluap1, a component of the IFT-B complex, and showed that cluap1 mutants have defects in ciliogenesis that result in photoreceptor degeneration (Lee et al., 2014). We performed a series of studies on the HLH protein, Id2a, and the gene regulatory network in which it operates. Id2a modulates retinoblast cell cycle kinetics and thereby influences eye growth, as well as neuron and glia formation in the retina (Uribe and Gross, 2010). Id2a acts upstream of the Notch pathway in regulating cell cycle progression and neuronal differentiation, and via RNA-Seq, we have identified numerous additional downstream Id2a effectors (Uribe et al., 2012). In collaboration with Peter Hitchcock (University of Michigan), we also showed that Id2a functions downstream of the secreted factor Midkine-a during retinoblast proliferation and differentiation (Luo et al., 2012). Finally, through a series of studies on the zebrafish patched1 mutant (formerly patched2) we identified roles for patched1 in regulating the size of the retinal progenitor pool in the CMZ (Bibliowicz and Gross, 2009), and in preventing ectopic proliferation and retinal dysplasia in the post-embryonic retina (Bibliowicz and Gross, 2011).
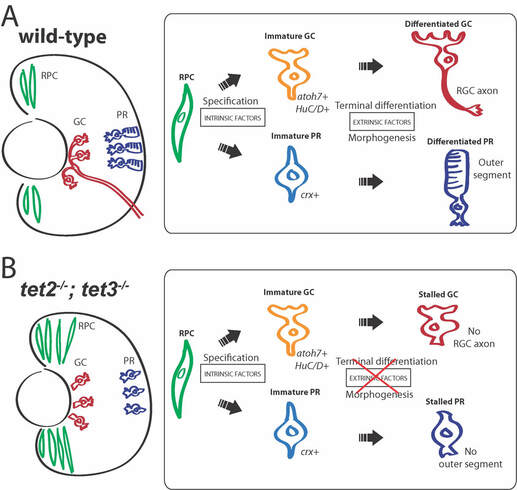
A major focus in our lab currently is on DNA methylation (5mC) and hydroxymethylation (5hmC) during eye development and, specifically, during the transition from retinal progenitor cell (RPC) to differentiated retinal neuron and in maintenance of retinal stem cells (RSCs). Our long-term goal is to generate genome-wide, single-nucleotide resolution 5mC and 5hmC profiles for RPCs of different competency states and for specific types of differentiated retinal neurons, and to correlate these profiles with quantitative gene expression data and then determine whether 5mC and 5hmC marks are required for normal gene expression and cell fate transitions in these cells. We hypothesize that epigenetic regulation of gene expression within RPCs and RSCs facilitates normal retinal development and growth by influencing the ability of these cells to remain as progenitors or to differentiate as retinal neurons. While a growing body of work demonstrates that epigenetic factors play key roles in retinal development, and possibly retinal disease, we do not yet understand how changes in DNA methylation or hydroxymethylation in RPCs and RSCs facilitate continued proliferation or differentiation, nor do we know whether these changes in methylation and hydroxymethylation are required for normal development or continued retinal growth and neurogenesis. While our genome data will be instrumental in predicting how epigenetic changes affect gene expression during RPC maturation and differentiation, and RSC maintenance, we need a mechanism to functionally test these predictions. Thus, we have systematically characterized and generated mutations in the genes that effect 5mC to 5hmC conversion (the tet2- and tet3 dioxygenases; Seritrakul and Gross, 2017), the maintenance methyltransferase, dnmt1 (Tittle et al., 2011; Angileri and Gross, 2020), and the genes encoding zebrafish de novo DNA methyltransferses, which methylate naked cytosines (the Dnmt3-family members; Seritrakul and Gross, 2014). We anticipate that the results of these studies will be of broad interest to the retinal development field, and translationally, for the continued development of stem cell-based therapies aimed at repairing or regenerating cells lost to disease or injury.
A major focus in our lab currently is on DNA methylation (5mC) and hydroxymethylation (5hmC) during eye development and, specifically, during the transition from retinal progenitor cell (RPC) to differentiated retinal neuron and in maintenance of retinal stem cells (RSCs). Our long-term goal is to generate genome-wide, single-nucleotide resolution 5mC and 5hmC profiles for RPCs of different competency states and for specific types of differentiated retinal neurons, and to correlate these profiles with quantitative gene expression data and then determine whether 5mC and 5hmC marks are required for normal gene expression and cell fate transitions in these cells. We hypothesize that epigenetic regulation of gene expression within RPCs and RSCs facilitates normal retinal development and growth by influencing the ability of these cells to remain as progenitors or to differentiate as retinal neurons. While a growing body of work demonstrates that epigenetic factors play key roles in retinal development, and possibly retinal disease, we do not yet understand how changes in DNA methylation or hydroxymethylation in RPCs and RSCs facilitate continued proliferation or differentiation, nor do we know whether these changes in methylation and hydroxymethylation are required for normal development or continued retinal growth and neurogenesis. While our genome data will be instrumental in predicting how epigenetic changes affect gene expression during RPC maturation and differentiation, and RSC maintenance, we need a mechanism to functionally test these predictions. Thus, we have systematically characterized and generated mutations in the genes that effect 5mC to 5hmC conversion (the tet2- and tet3 dioxygenases; Seritrakul and Gross, 2017), the maintenance methyltransferase, dnmt1 (Tittle et al., 2011; Angileri and Gross, 2020), and the genes encoding zebrafish de novo DNA methyltransferses, which methylate naked cytosines (the Dnmt3-family members; Seritrakul and Gross, 2014). We anticipate that the results of these studies will be of broad interest to the retinal development field, and translationally, for the continued development of stem cell-based therapies aimed at repairing or regenerating cells lost to disease or injury.
