Eye Evolution
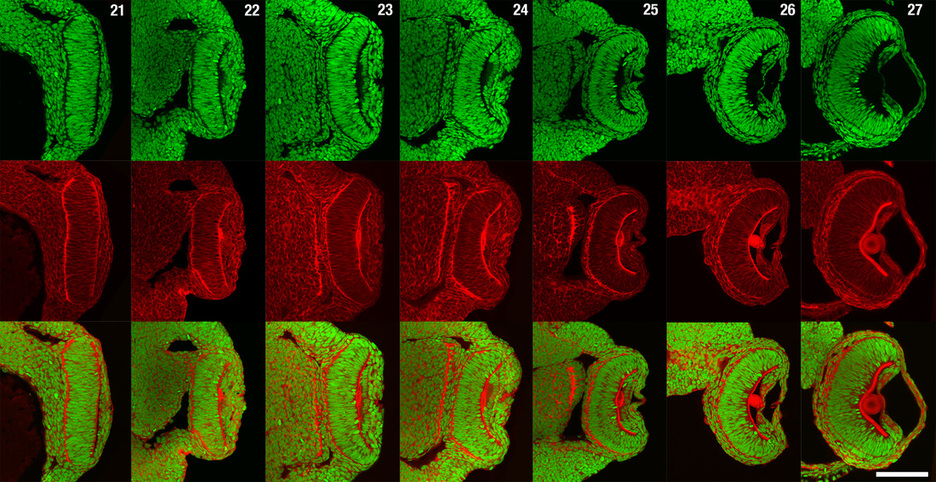
To better understand the eye of the Urbilaterian ancestor and how complex eye morphologies can evolve, it is essential to establish a Lophotrochozoan model for the development of a cephalic, complex image-forming eye. We have chosen to build off of the rich history of embryology and neurobiology using Loligo pealeii, and further develop it as a model Lophotrochozoan for studies of eye evolution (Koenig et al., 2016). Although Loligo pealeii has been the subject of developmental biology research for over 50 years, molecular analyses are in their infancy in the system. Research in the Cephalopod eye has mainly focused on mature tissues or on Pax-6 as a master regulator of eye specification. Gene products involved in patterning the developing eye and regulating morphogenesis remain almost entirely unknown. Our focus has been to develop Loligo pealeii as a system where developmental, molecular and evolutionary studies can be performed to elucidate the molecular basis of eye formation in the Lophotrochozoa, and by extension, across the Bilateria. To achieve this, we have generated a substantial amount of “infrastructure” for genomic and molecular studies, and are now using these data for more directed experiments on eye formation. For example, we generated a staging series that covers all stages of embryonic eye formation, a fate map of the early eye field, and we performed a series of detailed studies on cell proliferation, cell death, and retinal organization (Koenig et al., 2016). There are no published Cephalopod genomes, making molecular studies slow and tedious. To address this problem, we sequenced and assembled the Loligo pealeii transcriptome, generating over 100 gigabases of transcript sequence, which assembled into 72,591 isogroups with an N50 value of 1178 bp. This project has enabled us to make a substantial leap forward in beginning to identify the conserved regulatory network underlying eye development across the Bilateria. Indeed, we have taken both a candidate gene approach, having identified and characterized conserved regulators of eye development in Drosophila and vertebrates (e.g. the Pax6 cascade, Hh pathway, Notch pathway, and numerous other transcriptional regulators), and we have utilized RNA-Seq on dissected eyes and optic lobes from five stages of development to identify potentially novel regulators of eye development in Loligo pealeii. Finally, we are performing functional analyses of some of these conserved and novel factors, and our data support a role for the Notch pathway in regulating proliferation and differentiation in the Cephalopod retina (Koenig et al., 2016).
Ongoing work focuses on developing projects in other organisms where we can leverage their unique biology to uncover fundamental aspects of eye development and potentially develop these into new therapeutic approaches to treat human eye diseases (Koenig and Gross, 2020).
Ongoing work focuses on developing projects in other organisms where we can leverage their unique biology to uncover fundamental aspects of eye development and potentially develop these into new therapeutic approaches to treat human eye diseases (Koenig and Gross, 2020).
